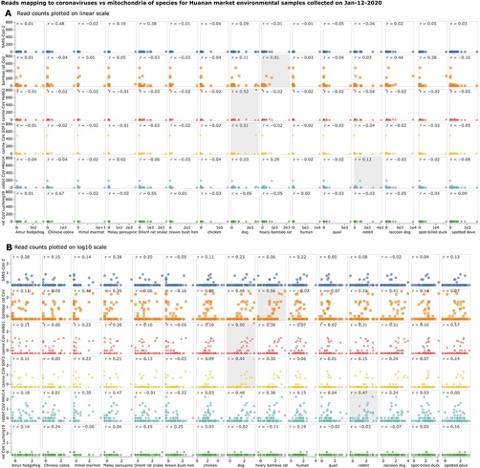
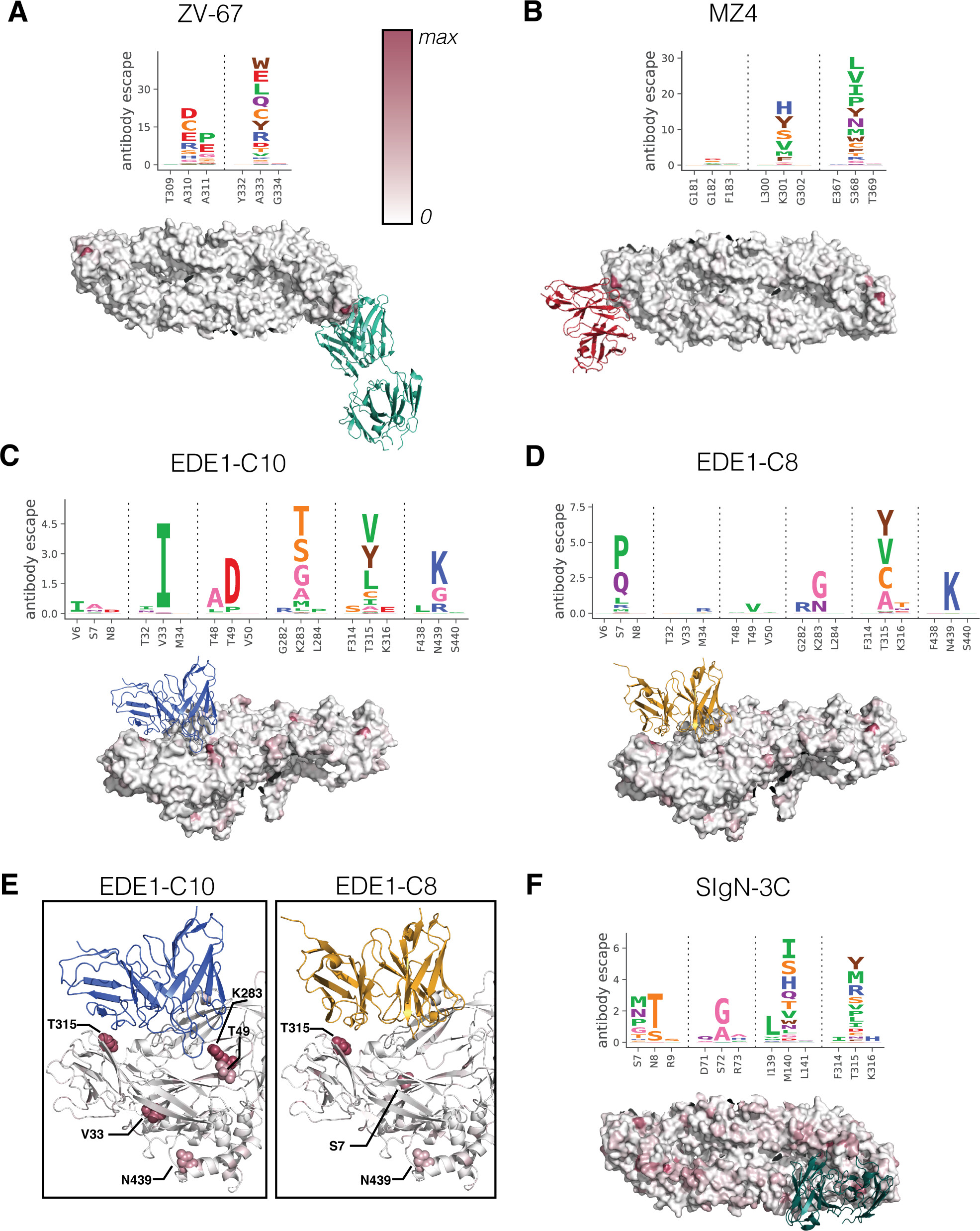
Papers
Papers from the Bloom Lab team. See also Google Scholar for a complete list of publications.
Filter by Keywords
Key Papers
These papers exemplify key current areas of research in the lab.
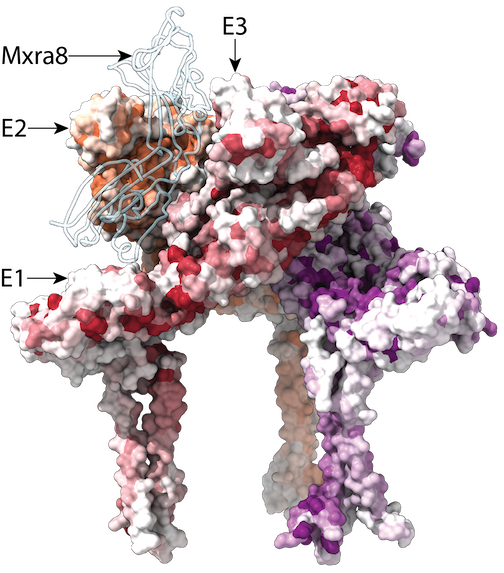
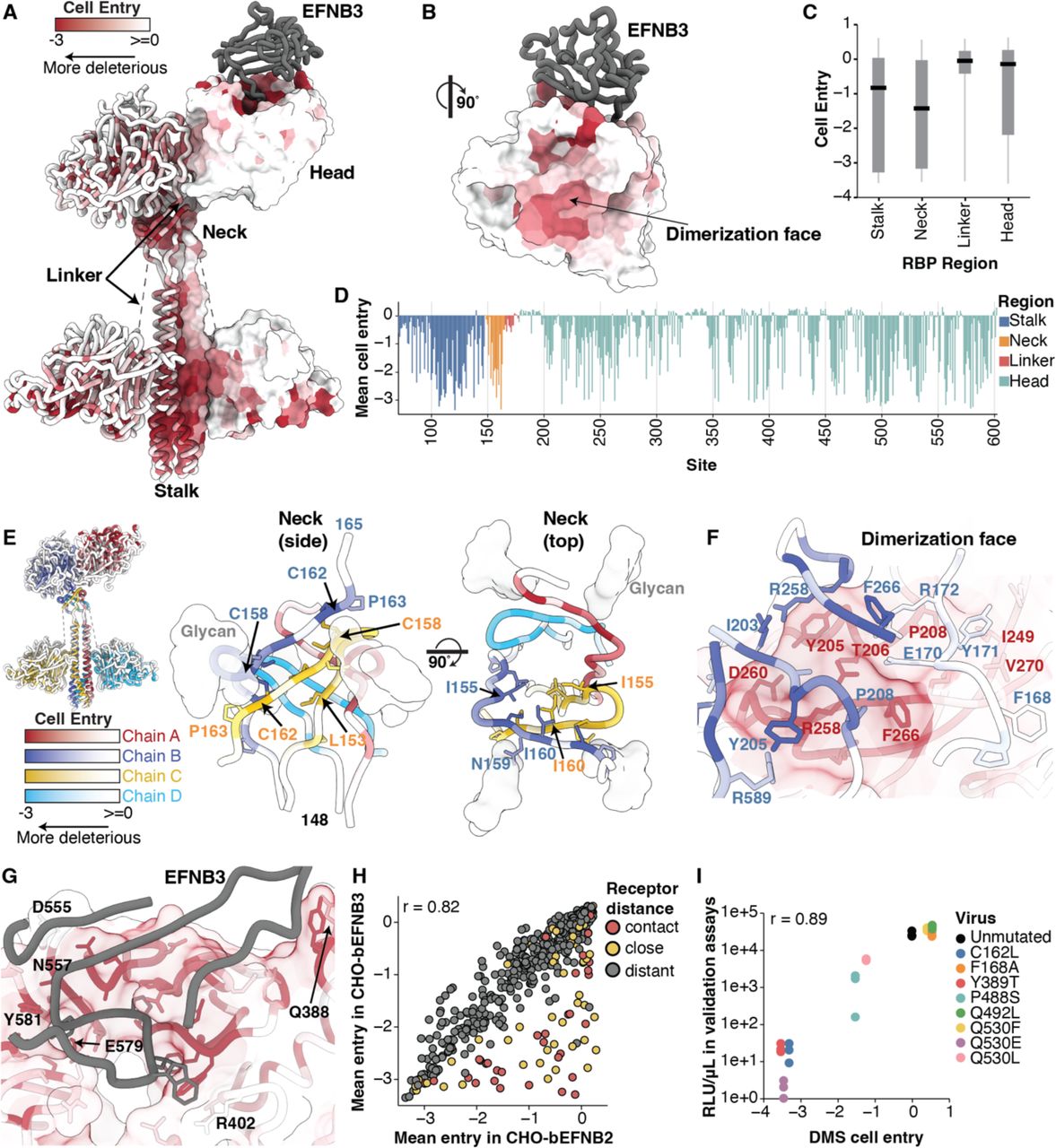
Determinants of human versus mosquito cell entry by the Chikungunya virus envelope proteins
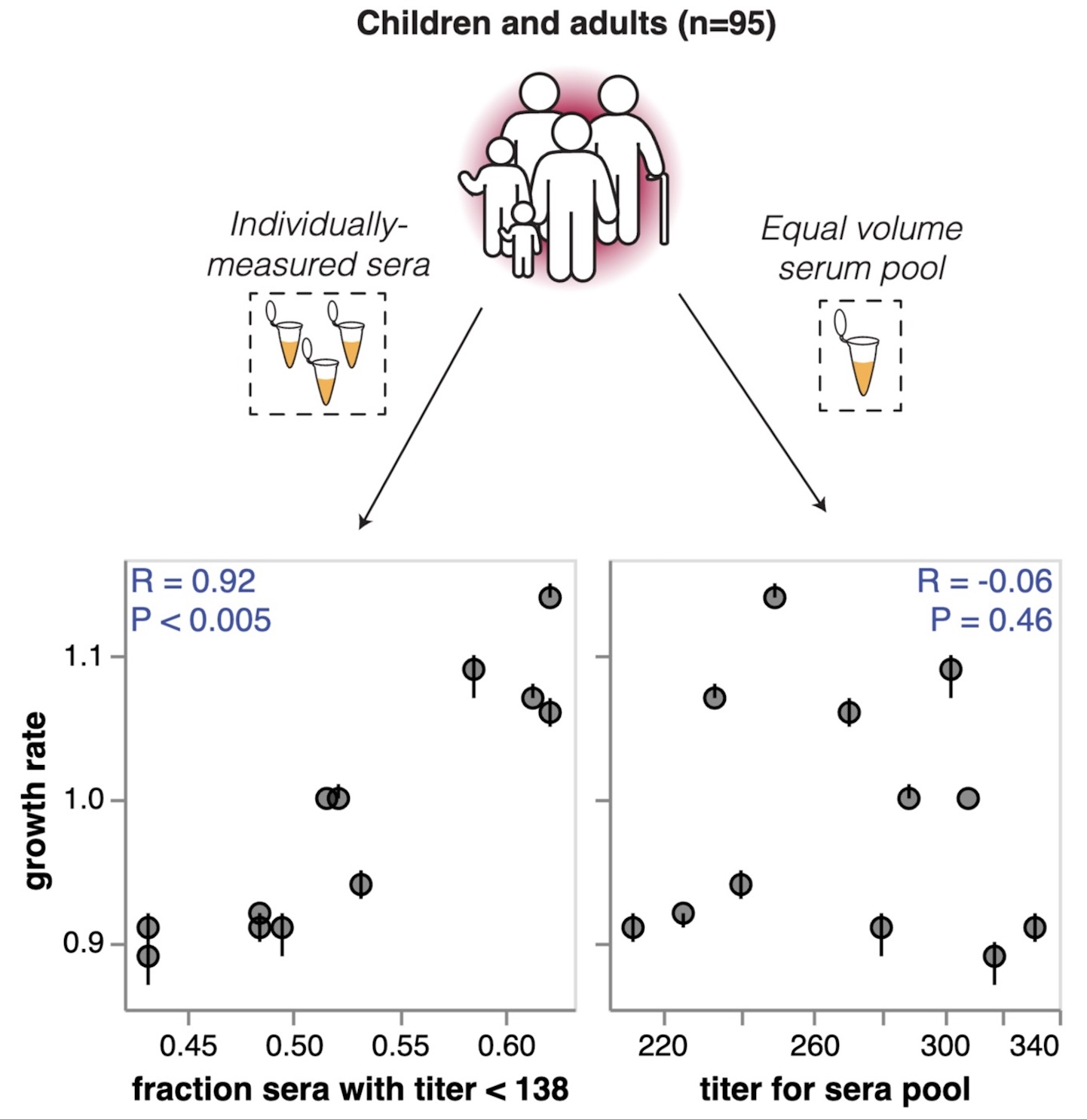
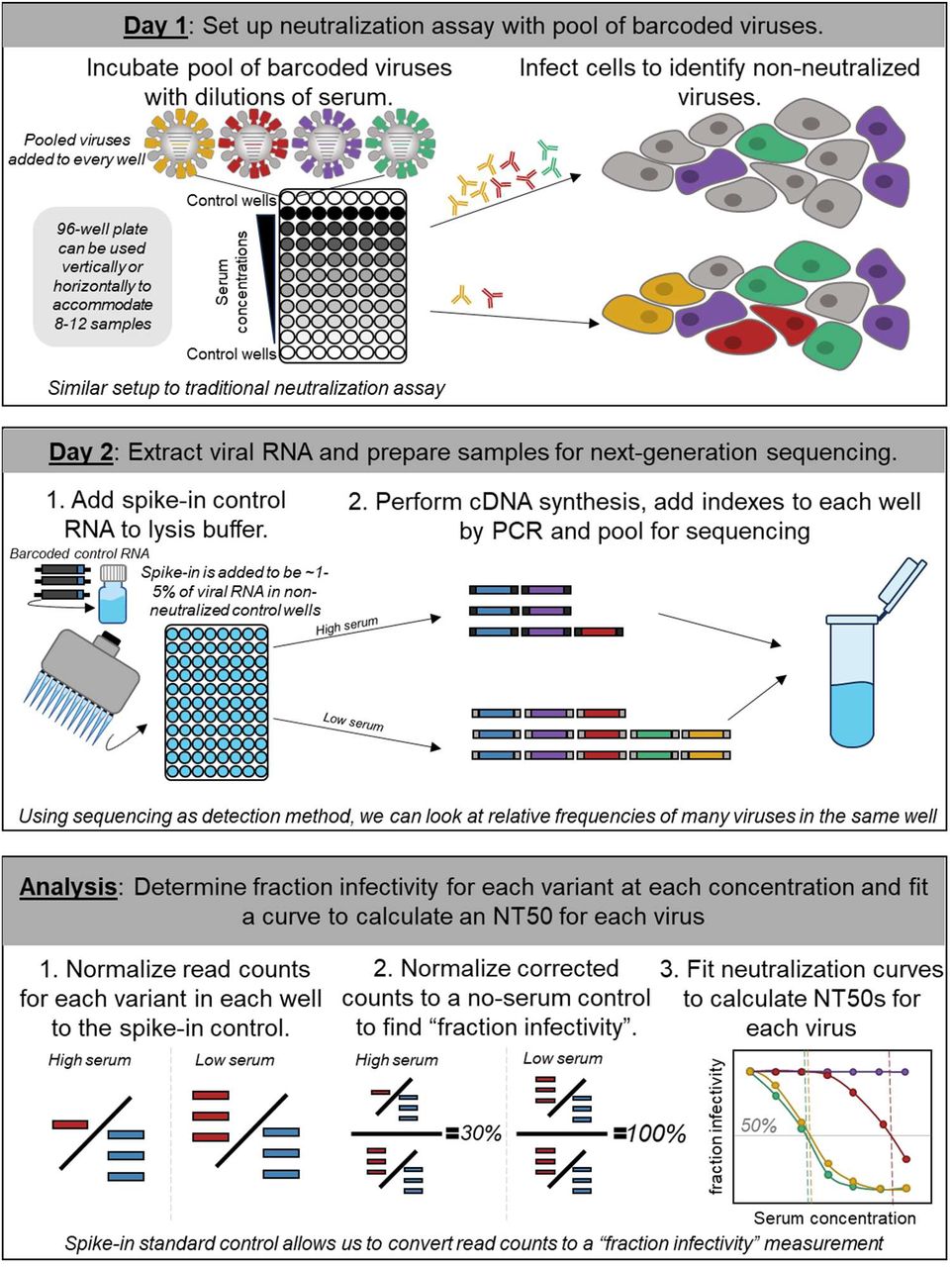
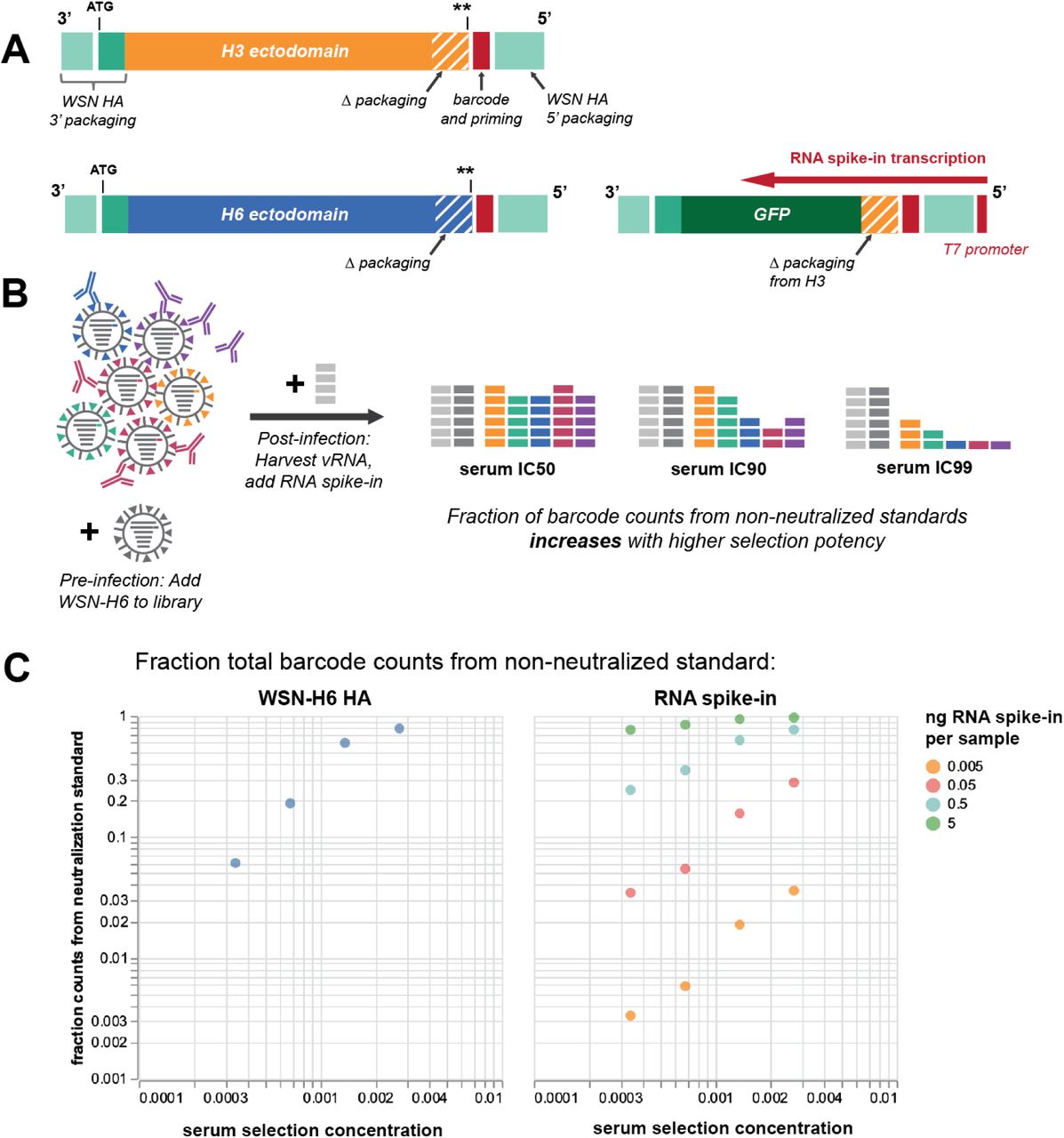
bioRxiv. 2025High-throughput neutralization measurements correlate strongly with evolutionary success of human influenza strains
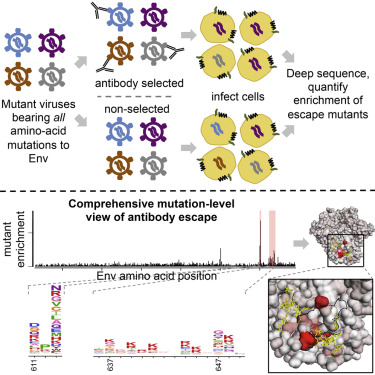
bioRxiv. 2025Deep mutational scanning of H5 hemagglutinin to inform influenza virus surveillance
PLoS Biology. 2024All Papers
Below is a complete list of primary research papers from our group.
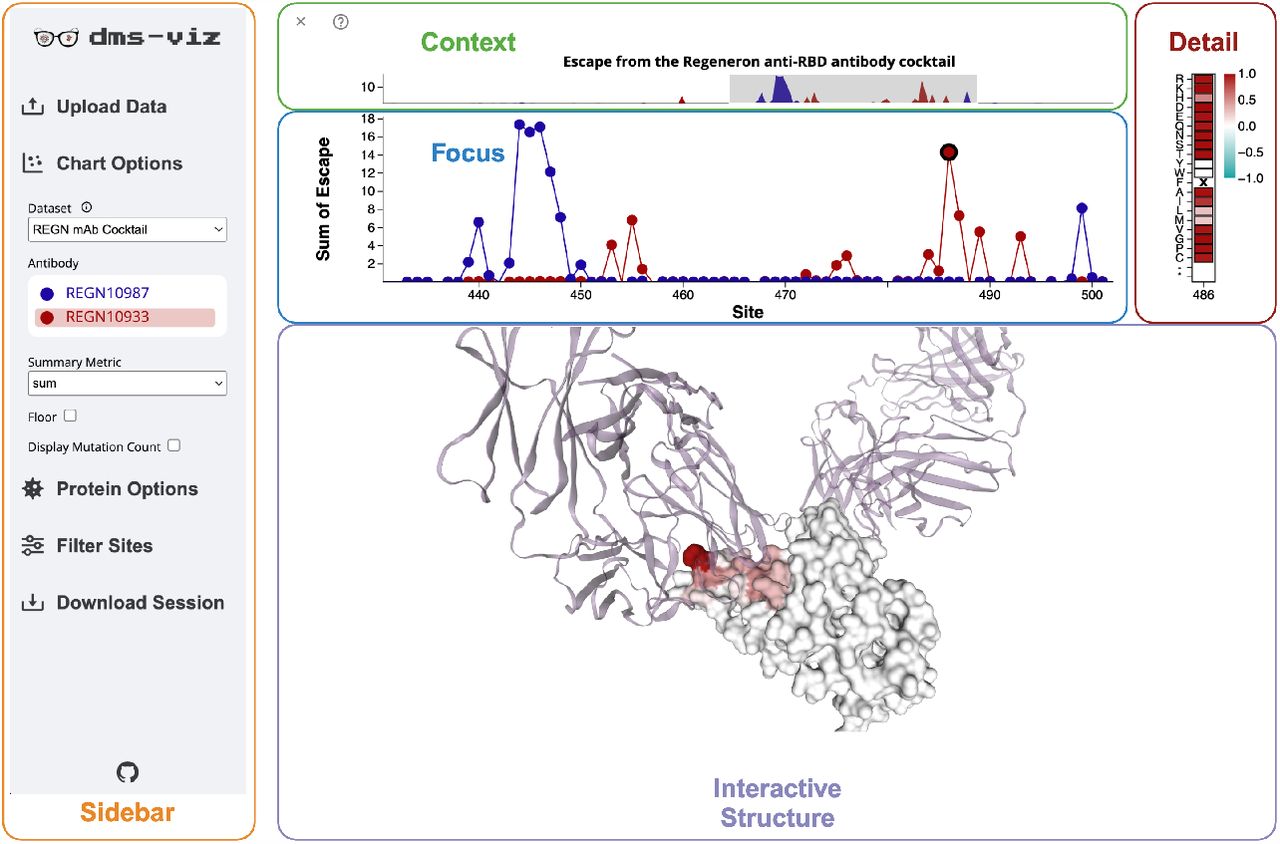
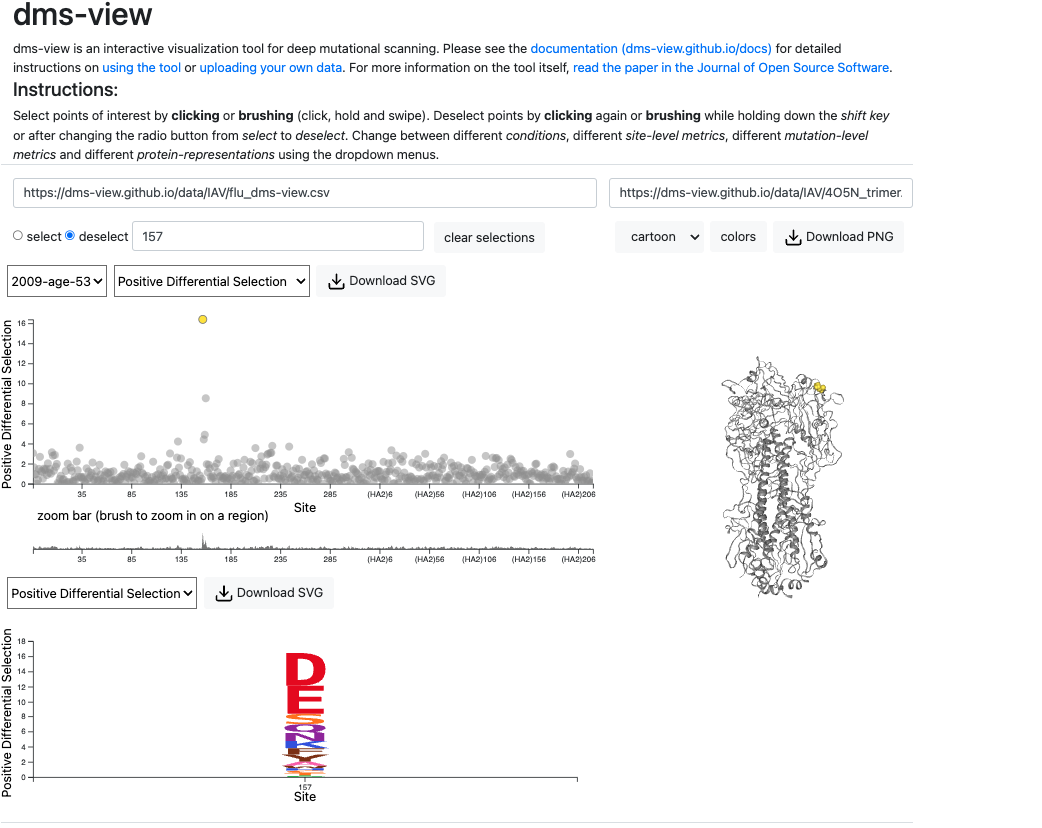
dms-viz: Structure-informed visualizations for deep mutational scanning and other mutation-based datasets
Journal of Open Source Software. 2024. doi:10.21105/joss.06129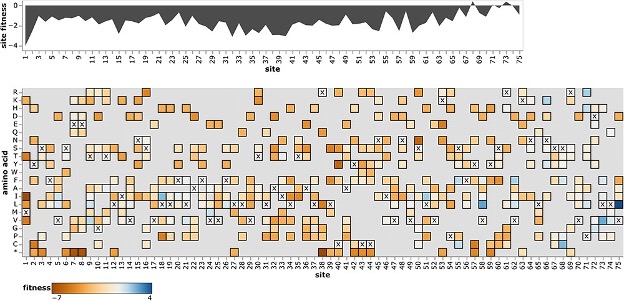
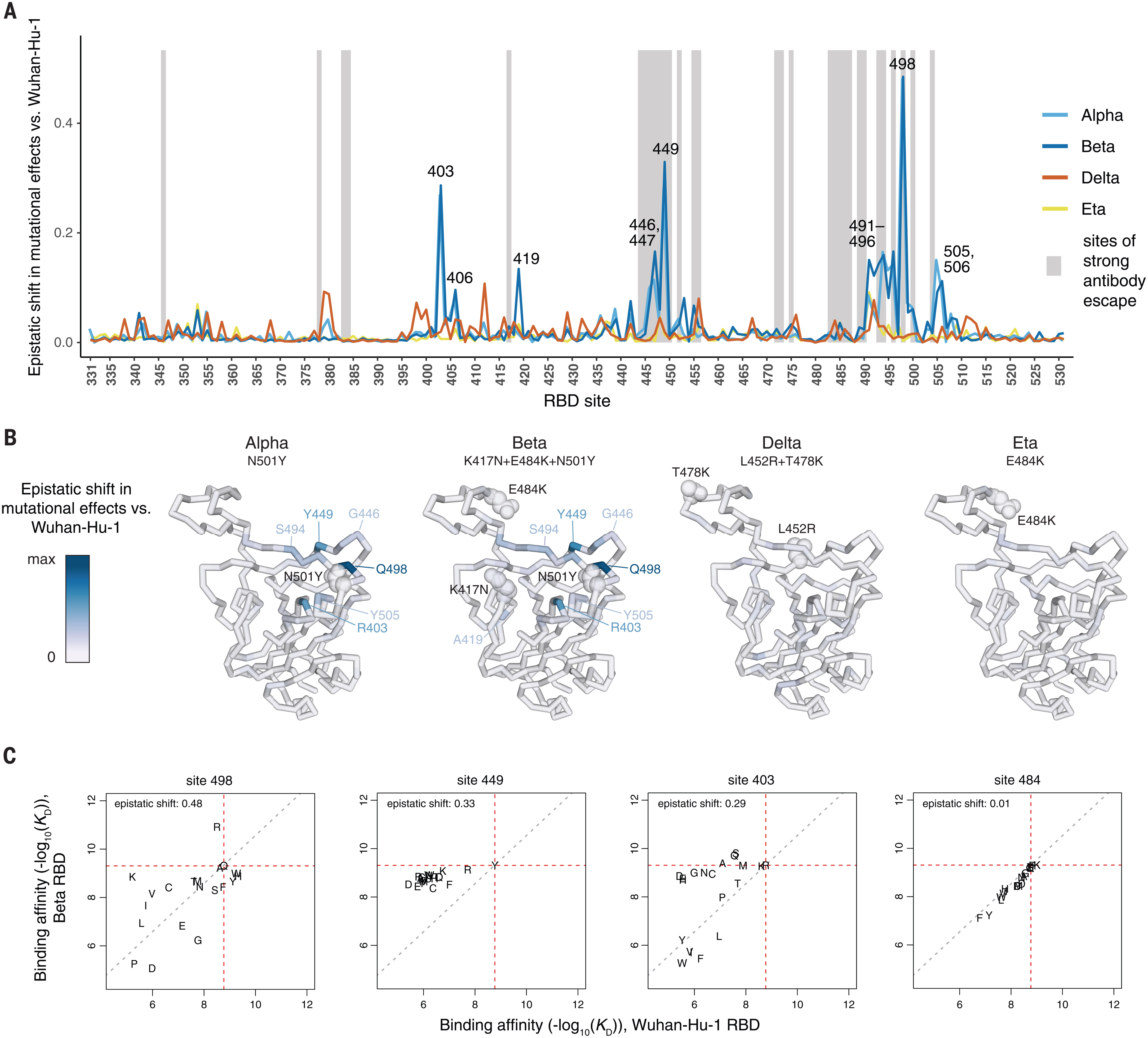
Fitness effects of mutations to SARS-CoV-2 proteins
Virus Evolution. 2023. doi:10.1093/ve/veae026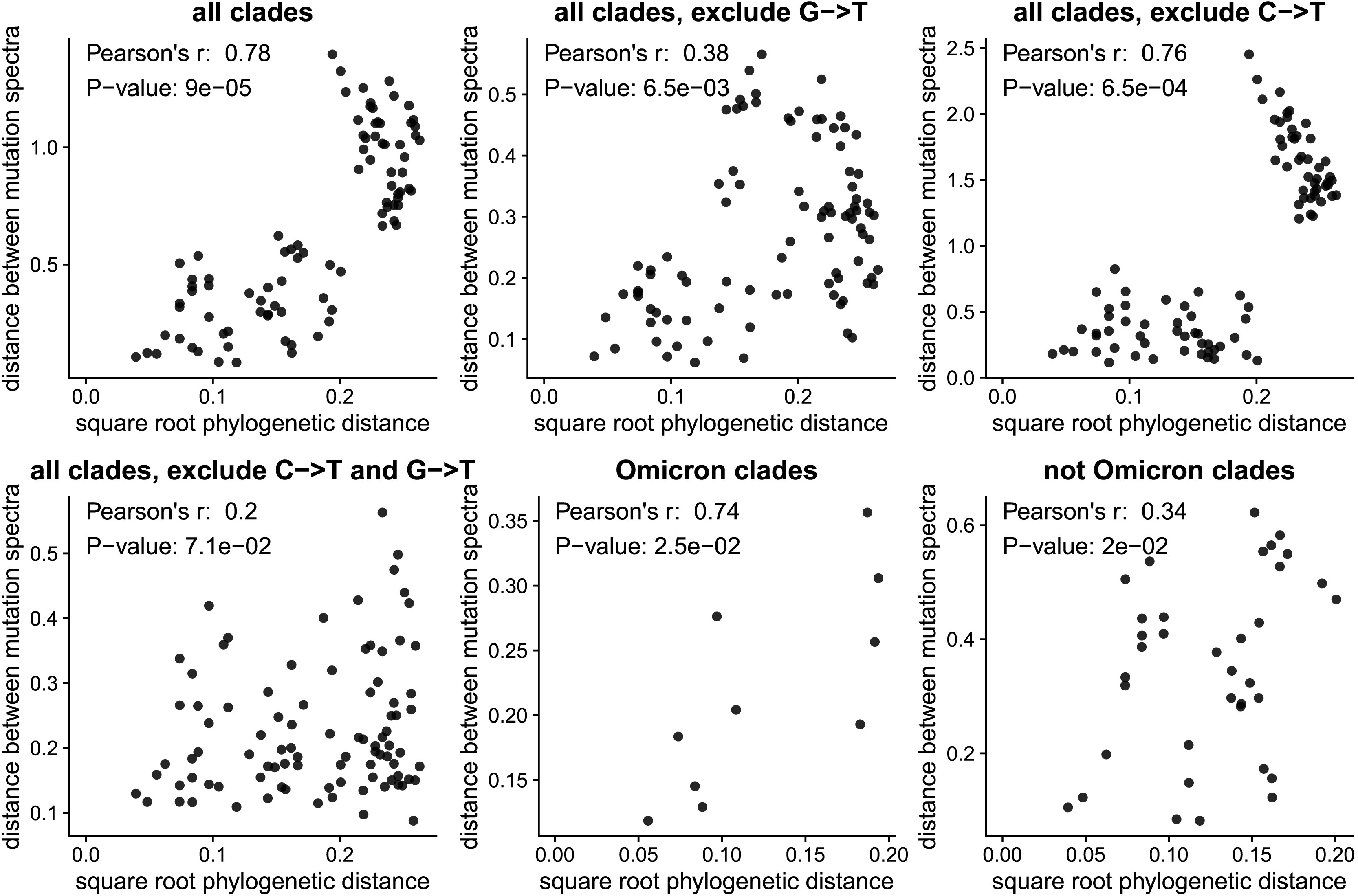
Evolution of the SARS-CoV-2 mutational spectrum
Molecular Biology and Evolution. 2023. doi:10.1093/molbev/msad085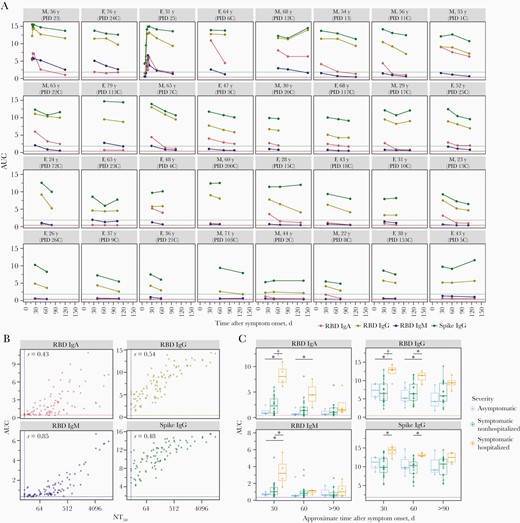
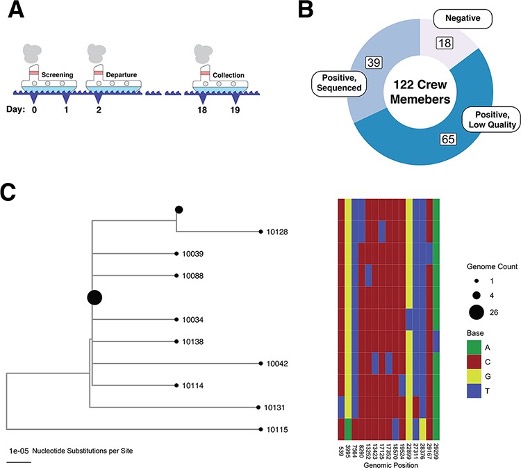
Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection
The Journal of infectious diseases. 2021. doi:10.1093/infdis/jiaa618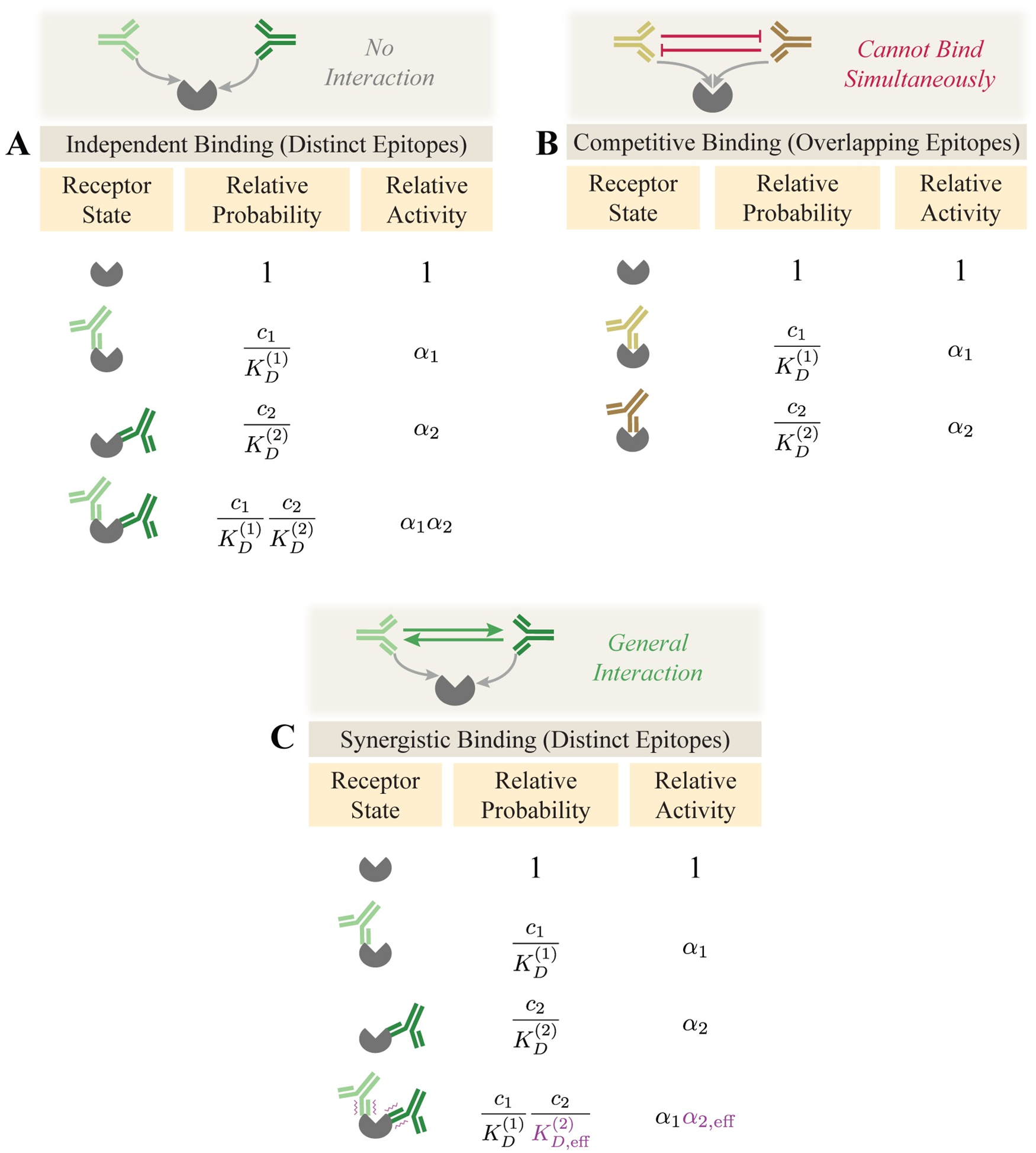
When two are better than one: Modeling the mechanisms of antibody mixtures
PLoS computational biology. 2020. doi:10.1371/journal.pcbi.1007830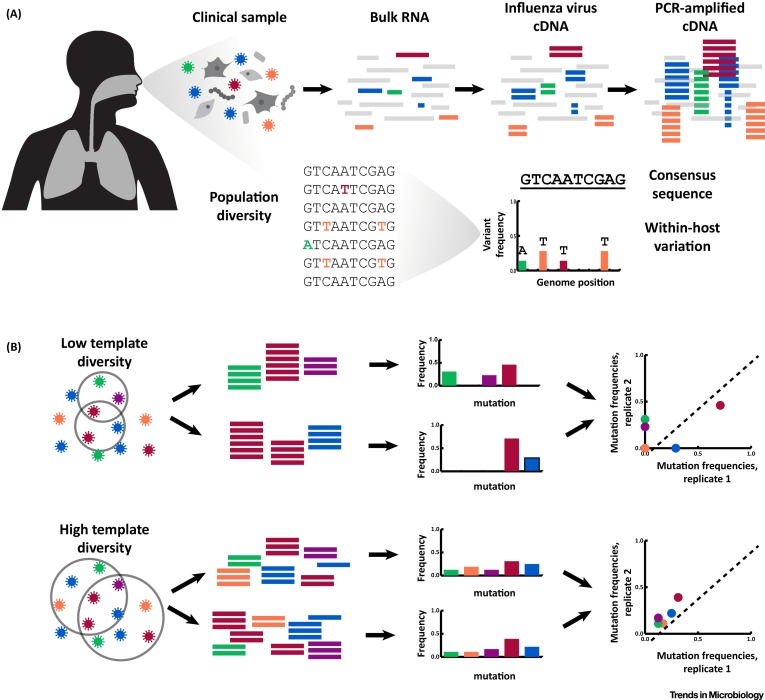
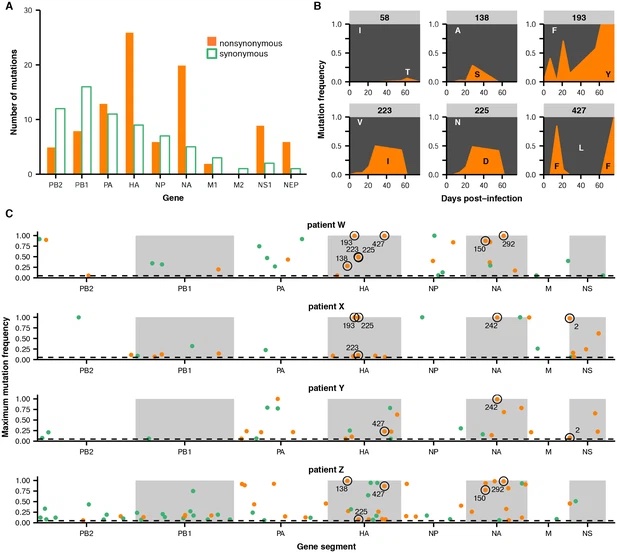
Within-host evolution of human influenza virus
Trends in Microbiology. 2018. doi:10.1016/j.tim.2018.02.007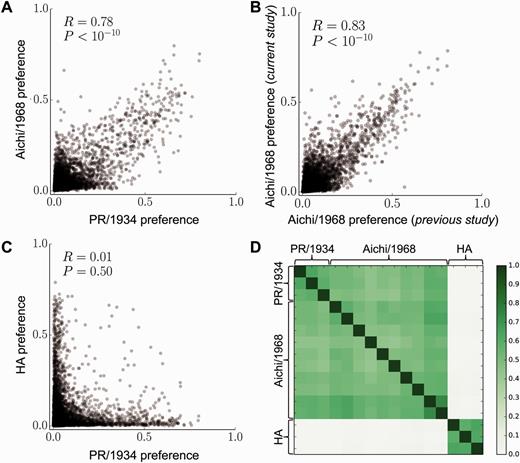
Site-specific amino acid preferences are mostly conserved in two closely related protein homologs
Molecular biology and evolution. 2015. doi:10.1093/molbev/msv167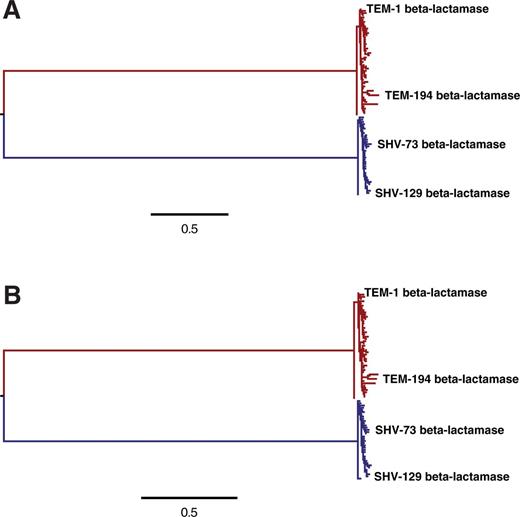
An experimentally informed evolutionary model improves phylogenetic fit to divergent lactamase homologs
Molecular biology and evolution. 2014. doi:10.1093/molbev/msu220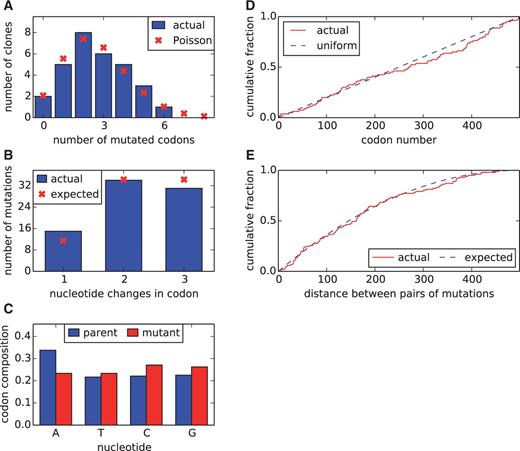
An experimentally determined evolutionary model dramatically improves phylogenetic fit
Molecular Biology and Evolution. 2014. doi:10.1093/molbev/msu173