Deep mutational scanning of rabies glycoprotein defines mutational constraint and antibody-escape mutations
In a new study we posted on bioRxiv today, we measured how all mutations to the rabies virus glycoprotein (G) affect cell entry and antibody neutralization. This study sheds light on mutational constraints on class III viral fusion proteins, suggests ways to stabilize G for vaccine antigens, and quantifies the robustness of antibodies (including ones being used or developed for clinical use) to rabies genetic variation.
Background
Rabies virus infects many different mammalian species. In humans, rabies is lethal without post-exposure prophylaxis (which can take the form of serum or antibody administration) or vaccination. Roughly 60,000 people die annualy from rabies infections, with the highest toll in Africa and Asia. Efforts are underway to make new vaccines and develop antibody treatments, such as Rabishield.
Vaccines and antibodies target G, which mediates viral receptor binding and cell entry. G is a class III fusion protein, which unlike the more heavily studies class I viral fusion proteins (eg, influenza HA or SARS-CoV-2 spike) undergo reversible conformational changes. However, this conformational flexibility can pose a challenge for vaccine design and antibody isolation: the most potent antibodies bind to prefusion G, but many antibodies elicited by native G bind to other conformations. As a result, one strategy for vaccine design is to stabilize the prefusion conformation of G.
Mutation effects on cell entry
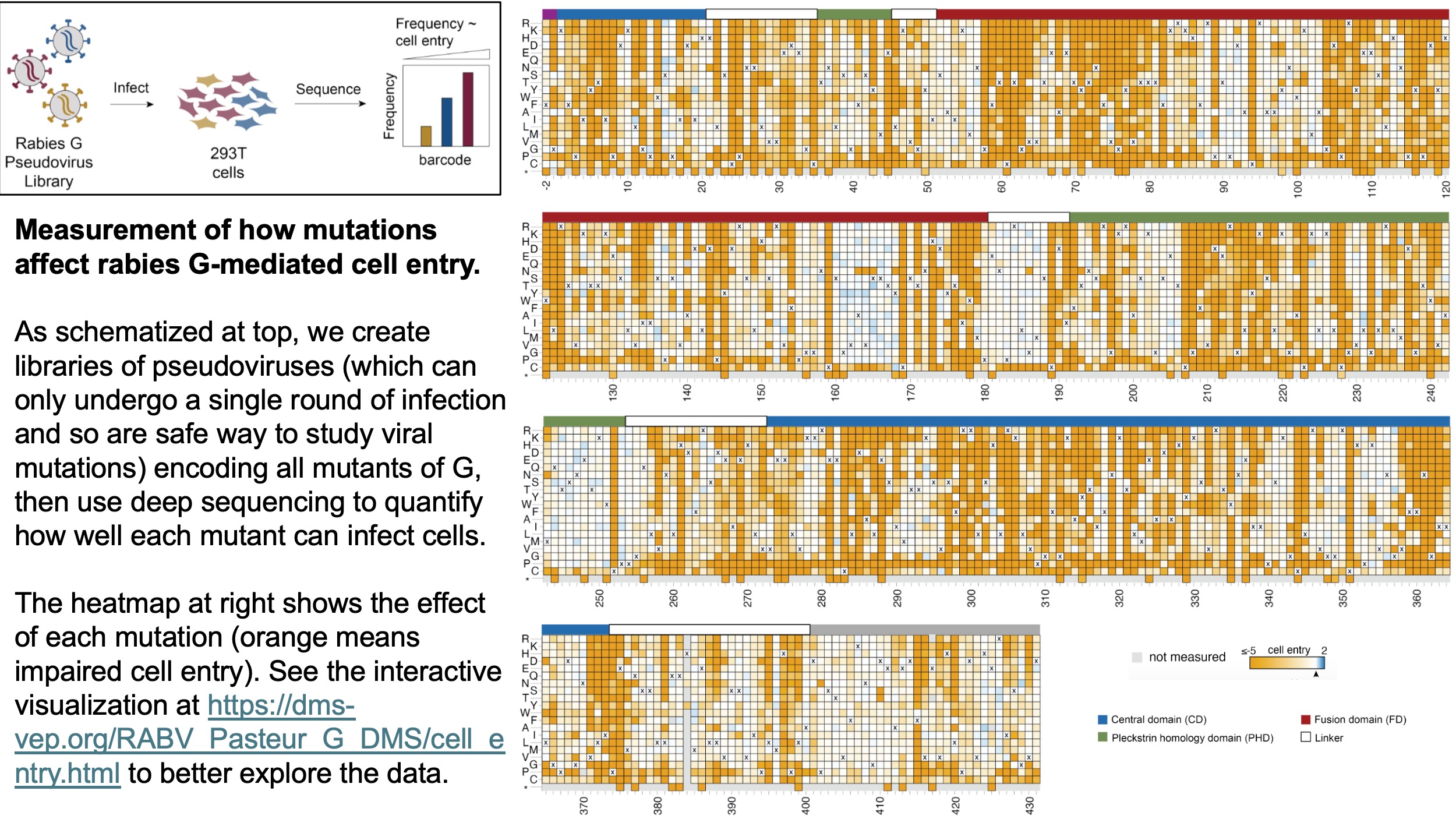
In our study, which was led by postdoc Arjun Aditham, we first measured how all G mutations affect cell entry using pseudoviruses, which are virions that can only undergo one round of infection and so offer a safe way to study viral mutations. The immediate result of these measurements is a huge heatmap of mutation effects, which is shown below but is best visualized interactively at https://dms-vep.org/RABV_Pasteur_G_DMS/cell_entry.html.
The mutational constraint can also be mapped on G's structure. Below is an image of the per-site constraint on the pre-fusion structure of G; see here for interactive visualizations on either the prefusion or extended intermediate conformations.
As described in the paper, analyses of the mutational effects on cell entry in the context of G's structure identifies mutations that are good candidates for stabilizing vaccine antigens.
Mutation effects on antibody escape
We next measured how all G mutations affect neutralization by eight monoclonal antibodies. Below is the escape map for 17C7, which is the antibody in Rabishield (see here for an interactive version of the 17C7 escape map).
These high-throughput measurements of how all G mutations affect antibody neutralization correlate exceedingly well with traditional pseudovirus neutralization assays:
We generated similar escape maps for a total of eight monoclonal antibodies that neutralize rabies; these maps can be viewed interactively as heatmaps or on the G structure at https://dms-vep.org/RABV_Pasteur_G_DMS/escape.html.
Antibody escape among natural rabies strains
Given that rabies is genetically diverse, a crucial question for antibody development is whether there are natural strains that escape any given antibody. We first examined the frequency of escape mutations as measured in our deep mutational scanning for each antibody against all natural strains. As shown below (and interactively here), escape mutations from all antibodies are present in natural strains, but their prevalence differs among antibodies.
We used the deep mutational scanning to predict escape from each antibody for all natural strains under an additive model of mutation effects. Below is a phylogeny colored by predicted escape from antibody 17C7 (see https://nextstrain.org/groups/jbloomlab/dms/rabies-G for interactive Nextstrain trees for all antibodies). As can be seen in the image below, we validated several of the strains predicted to escape this antibody actually did.
Interactive results and thanks
The results are available for further interactive exploration at https://dms-vep.org/RABV_Pasteur_G_DMS.
The study was led by Arjun Aditham in our lab, with help from Caelan Radford and Caleb Carr in our group, as well as Naveen Jasti and Neil King at the Institute for Protein Design.