SARS-CoV-2 neutralizing antibody specificities differ dramatically between recently infected infants and immune-imprinted individuals
In a new study that we posted on bioRxiv today, we showed that there are striking differences in how serum neutralizing antibodies target the SARS-CoV-2 spike depending on a person's exposure history. This result suggests that growing population immune heterogeneity is likely becoming of growing importance in shaping SARS-CoV-2 evolution.
Background
A person's immune response to a virus is "imprinted" by the first exposure, with subsequent exposure to new variants of the virus primarily activating pre-existing B cells. This imprinting has been shown to play an important role in shaping antibody repertoires to SARS-CoV-2.
Most of the global population was imprinted by the spike of an early SARS-CoV-2 strain: either they were vaccinated with the original COVID-19 vaccine that contained the Wuhan-Hu-1 strain, or they were infected in 2020 or 2021 by a similar viral variant. However, a small but growing fraction of the population has instead been imprinted by a more recent viral strain. This fraction of the population consists largely of infants who were first exposed to later viral variants.
In our study, we worked with the groups of Mary Staat (Cincinnati Children's Hospital) and Helen Chu (University of Washington) to compare the polyclonal serum neutralizing antibodies in adults that had been imprinted by an early SARS-CoV-2 strain and then infected with a XBB* variant in 2023, versus infants who had only been infected with a XBB* variant in 2023.
Dramatic differences in specificity of serum neutralization between imprinted adults/children and infants with just a single XBB* infection
Bernadeta Dadonaite in our group compared these sera by using deep mutational scanning with pseudoviruses expressing the XBB.1.5 spike to examine which mutations reduced neutralization be each set of sera. She had previously shown that for imprinted adults, mutations at some key sites in the spike receptor-binding domain (RBD) caused the most sera escape. In this new study, she found the same thing was true for imprinted children.
But when we performed the deep mutational scanning of the sera from the infants only infected by XBB*, we were stunned by what we found! The sites of strongest escape from the infant sera were not even in the RBD, but instead were in the spike N-terminal domain (NTD), as shown below:
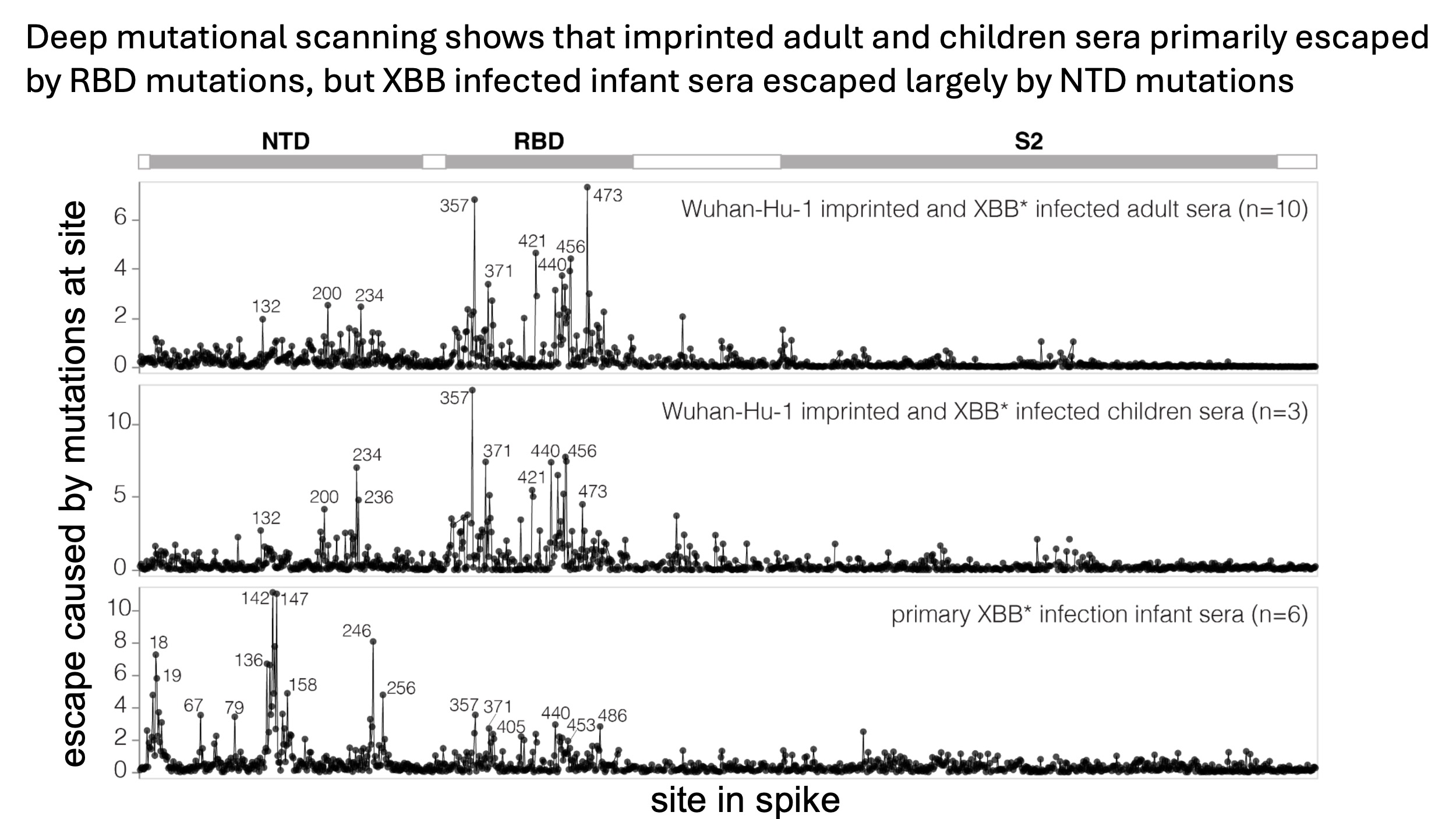
As described in the full paper, this variation between the different sets of sera is in contrast to the differences between sera within the same set, where there is some variation but mostly sites of escape are roughly similar.
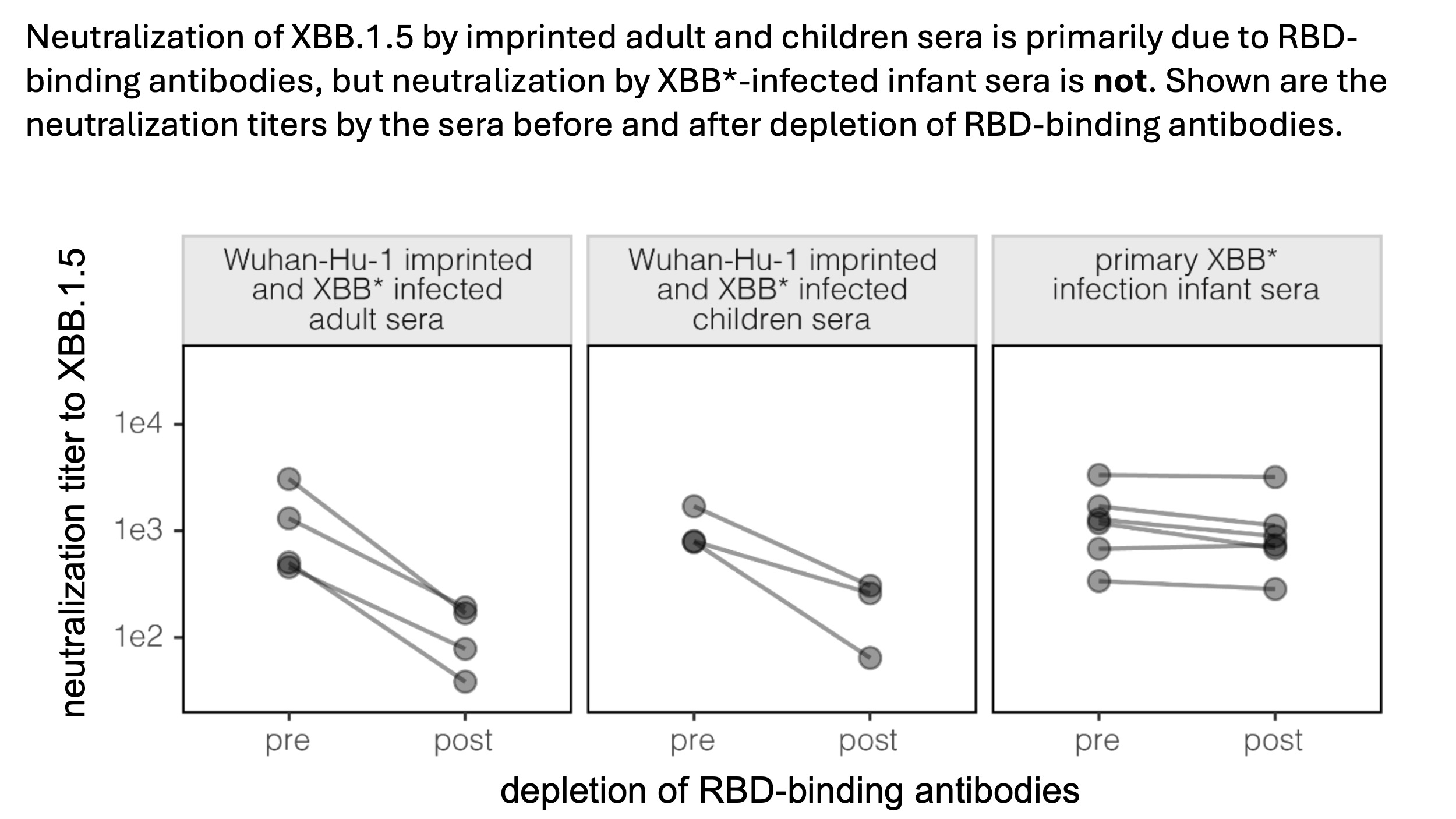
To validate the deep mutational scanning, we measured neutralization of XBB.1.5 spike-pseudovirus before and after depleting RBD-binding antibodies. Consistent with the deep mutational scanning, depleting RBD-binding antibodies greatly reduced neutralization by imprinted adult/children sera, but not XBB* infected infant sera.
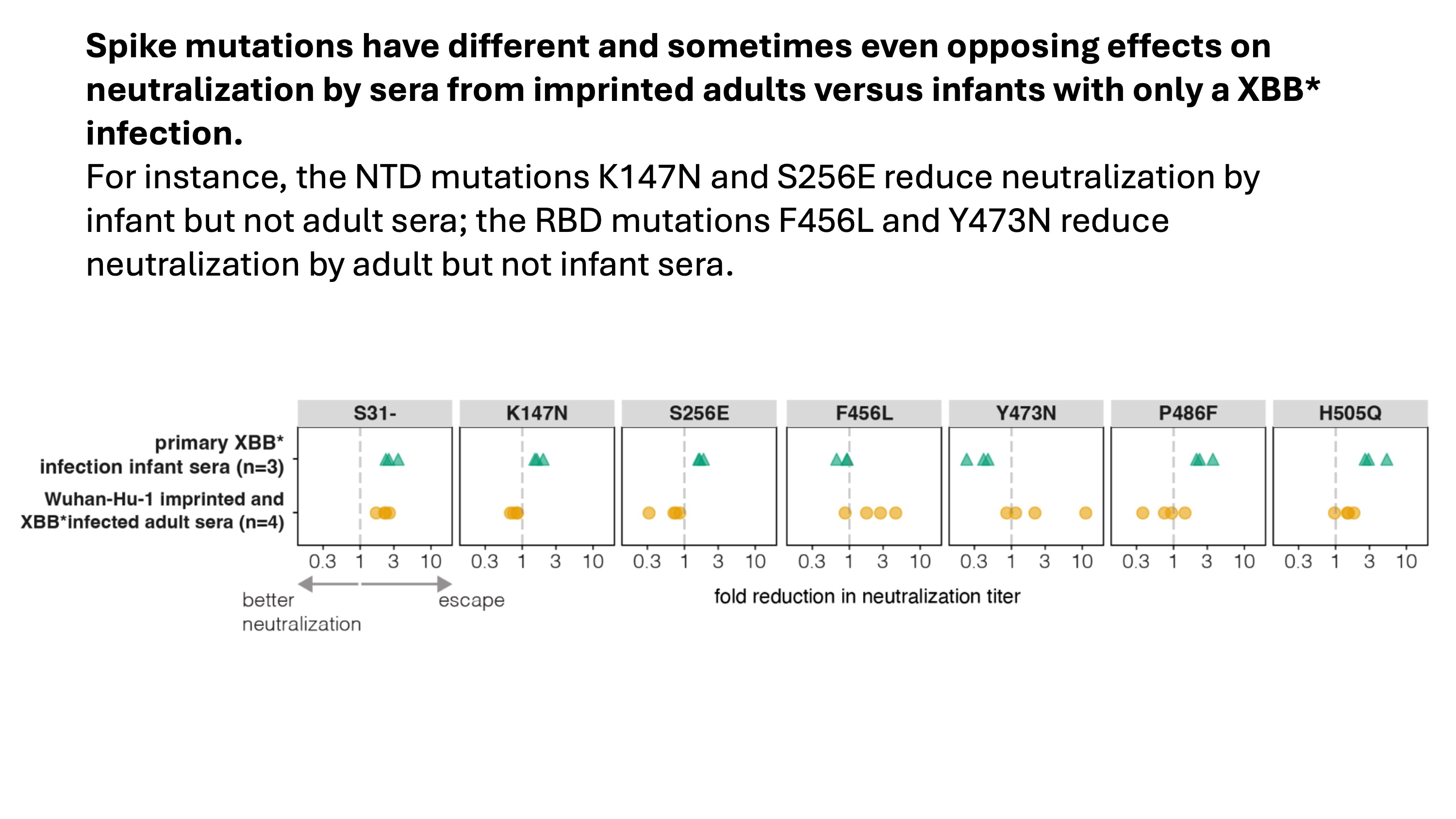
These results imply SARS-CoV-2 mutations can have very different impacts on serum antibody neutralization depending on a person's exposure history. We used neutralization assays to confirm this fact for a subset of mutations as shown below.
Vaccination boosts RBD targeting for infants
Subsequent vaccination of the XBB* infected infants does increase the RBD-directed neutralization of their sera, such that it strongly targets both the NTD and RBD. The reason is unclear, but we speculate it might be affinity maturation of potent RBD antibodies.
Implications for viral evolution
Overall, we think this heterogeneity among people could have substantial impacts on viral evolution. The evolutionary selection pressures on the virus become more complex if the same mutation only confers an antigenic benefit in a fraction of infected people.
Most prior studies of SARS-CoV-2 antigenic evolution have used sera mostly from imprinted adults. Yet the recent evolution of the virus has also seen selection for NTD mutations. Indeed, over the last five years spike mutations have been strongly selected in both RBD and NTD neutralizing antibody epitopes as shown below:
What causes the differences between imprinted individuals and singly infected infants?
We don't know the answer for certain! But read the discussion of the full paper for our speculation.
Interactive results and thanks
The results are available for further interactive exploration at https://dms-vep.org/SARS-CoV-2_XBB.1.5_spike_DMS_infant_sera/.
The study was led by Bernadeta Dadonaite in our lab, and made possible by the invaluable human cohorts maintained by our collaborators and co-authors:
IMPRINT study (Cincinnati Children's Hospital):
- Mary Staat
- Allison Burrell
- David Haslam
- Daniel Payne
HAARVI study (University of Washington):
- Helen Chu
- Jenni Logue