Our Research
Deep Mutational Scanning
Our lab uses deep mutational scanning to experimentally measure how tens-of-thousands of mutations to viral proteins affect key properties including function, immune escape, and receptor binding.
We primarily perform these experiments using a pseudovirus system that allows us to safely characterize mutants of entry proteins from a wide range of viruses, including SARS-CoV-2 spike, influenza hemagglutinin, Lassa virus GPC, HIV envelope, and Nipah virus RBP.
Deep mutational scanning can inform efforts to forecast the evolution of human seasonal viruses and surveil the evolution of potential pandemic viruses. To facilitate the use of deep mutational scanning for these important goals, we develop interactive visualization tools and data analysis pipelines. See here for an example of how we analyze and visualize large datasets to inform the study of viral evolution.
Interplay of Immunity and Viral Evolution
We study immunity and viral evolution at both the population and single-cell levels.
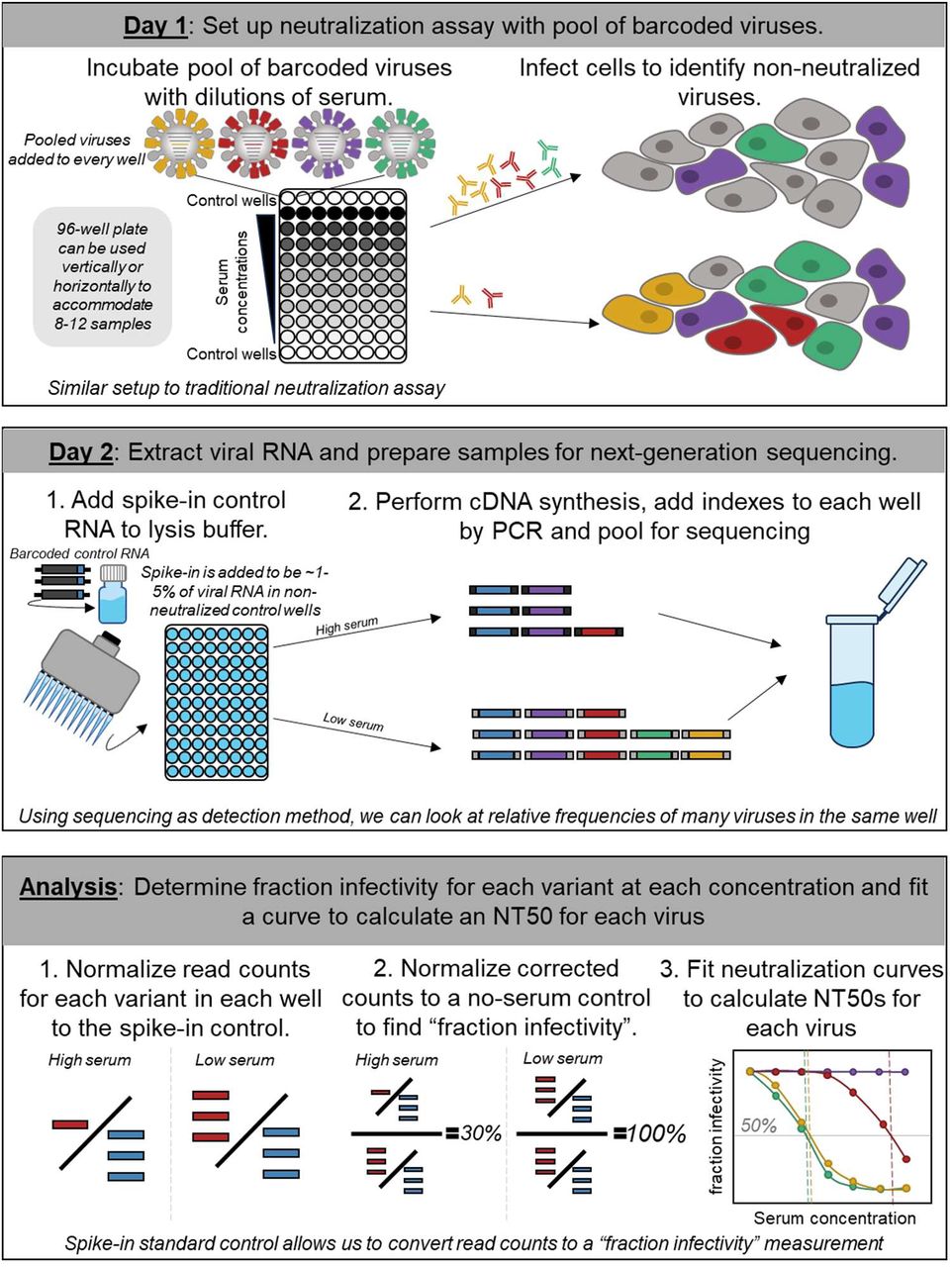
At the population level, differences in exposure history and immune imprinting lead human individuals to make antibody responses that target different regions of rapidly evolving viruses like influenza and SARS-CoV-2. This population heterogeneity has profound implications for viral evolution and disease susceptibility, as it causes viral mutations to impact the immunity of different individuals differently. We are characterizing this population heterogeneity using both deep mutational scanning and a sequencing based-neutralization assay we developed that increases the throughput of traditional neutralization assays by several orders of magnitude (see schematic at left).
At the single-cell level, we developed approaches to sequence viruses in single cells and quantify how many progeny each infected cell produces. We use these approaches to understand how viral variation impacts the outcome of infection in individual cells.
Big Datasets and Viral Evolution
We also develop new ways to leverage large datasets to better understand viral evolution.
We have come up with a way to leverage the millions of publicly available SARS-CoV-2 sequences to estimate the effect of individual mutations on viral fitness (see this paper and these slides). We've also created a platform to visualize the mutational effects to aid in interpretation of viral evolution.
We have also integrated thousands of deep mutational scanning measurements into an antibody-escape calculator that was widely used during the SARS-CoV-2 pandemic to understand the antigenic effects of viral mutations.
We also have projects that involve analyzing the evolution of viruses within individual infected humans, and developing models to understand epistasis among viral mutations.