Formatting CodonVariantTable plots¶
A CodonVariantTable generates informative plots about a deep mutational scanning experiment. Here are some tips on how to format those plots.
Setup for notebook¶
Import Python modules / packages:
[1]:
import random
import tempfile
import warnings
from IPython.display import display, Image
import numpy
from plotnine import *
import dms_variants.codonvarianttable
import dms_variants.plotnine_themes
import dms_variants.simulate
from dms_variants.constants import CBPALETTE, CODONS_NOSTOP
Hide warnings that clutter output:
[2]:
warnings.simplefilter("ignore")
Simulate a CodonVariantTable¶
We simulate a CodonVariantTable to use to demonstrate the plot formatting. Set parameters that define the simulated data:
[3]:
seed = 1 # random number seed
genelength = 40 # gene length in codons
libs = ["lib_1", "lib_2"] # distinct libraries of gene
variants_per_lib = 500 * genelength # variants per library
avgmuts = 2.0 # average codon mutations per variant
bclen = 16 # length of nucleotide barcode for each variant
variant_error_rate = 0.01 # rate at which variant sequence mis-called
avgdepth_per_variant = 200 # average per-variant sequencing depth
lib_uniformity = 5 # uniformity of library pre-selection
noise = 0.02 # random noise in selections
bottlenecks = { # bottlenecks from pre- to post-selection
"tight_bottle": variants_per_lib * 5,
"loose_bottle": variants_per_lib * 100,
}
Seed random number generator for reproducible output:
[4]:
random.seed(seed)
Simulate wildtype gene sequence:
[5]:
geneseq = "".join(random.choices(CODONS_NOSTOP, k=genelength))
print(f"Wildtype gene of {genelength} codons:\n{geneseq}")
Wildtype gene of 40 codons:
AGATCCGTGATTCTGCGTGCTTACACCAACTCACGGGTGAAACGTGTAATCTTATGCAACAACGACTTACCTATCCGCAACATCCGGCTGATGATGATCCTACACAACTCCGACGCTAGT
Generate a CodonVariantTable using simulate_CodonVariantTable function:
[6]:
variants = dms_variants.simulate.simulate_CodonVariantTable(
geneseq=geneseq,
bclen=bclen,
library_specs={
lib: {"avgmuts": avgmuts, "nvariants": variants_per_lib} for lib in libs
},
seed=seed,
)
Simulate counts for samples. First, we need a “phenotype” function to simulate the counts for each variant. We define this function using a SigmoidPhenotypeSimulator:
[7]:
phenosimulator = dms_variants.simulate.SigmoidPhenotypeSimulator(geneseq, seed=seed)
We then use the simulator to simulate some sample counts:
[8]:
counts = dms_variants.simulate.simulateSampleCounts(
variants=variants,
phenotype_func=phenosimulator.observedEnrichment,
variant_error_rate=variant_error_rate,
pre_sample={
"total_count": variants_per_lib * numpy.random.poisson(avgdepth_per_variant),
"uniformity": lib_uniformity,
},
pre_sample_name="pre-selection",
post_samples={
name: {
"noise": noise,
"total_count": variants_per_lib
* numpy.random.poisson(avgdepth_per_variant),
"bottleneck": bottle,
}
for name, bottle in bottlenecks.items()
},
seed=seed,
)
Add these counts to the CodonVariantTable:
[9]:
variants.add_sample_counts_df(counts)
Now we’ve completed the simulation of the CodonVariantTable:
Formatting plots¶
The plots returned by a CodonVariantTable above are all plotnine ggplot objects. So you can format them differently by setting a plotnine theme.
First make a plot using the default plotnine theme:
[10]:
# NBVAL_IGNORE_OUTPUT
p = variants.plotNumCodonMutsByType("all", samples=None)
_ = p.draw(show=True)
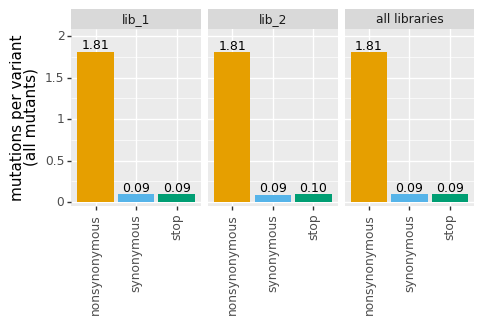
The dms_variants package defines a gray grid plotnine theme in dms_variants.plotnine_themes that gives an especially nice appearance for the plots. Here we set that theme and then re-draw the above plot:
[11]:
_ = theme_set(dms_variants.plotnine_themes.theme_graygrid())
[12]:
# NBVAL_IGNORE_OUTPUT
p = variants.plotNumCodonMutsByType("all", samples=None)
_ = p.draw(show=True)
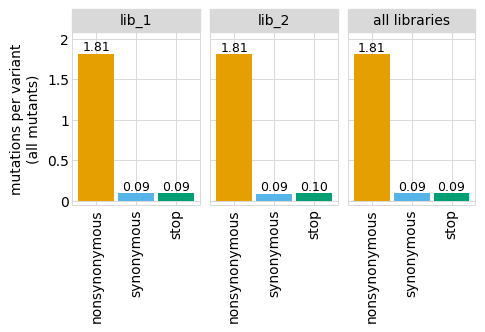
The plot looks even cleaner if we get rid of the vertical grid lines:
[13]:
# NBVAL_IGNORE_OUTPUT
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
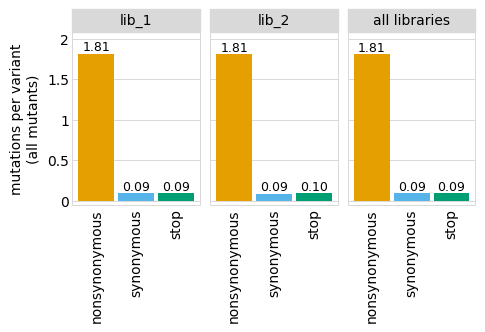
There are also lots of other themes defined by plotnine:
[14]:
# NBVAL_IGNORE_OUTPUT
theme_set(theme_bw())
p = variants.plotNumCodonMutsByType("all", samples=None)
_ = p.draw(show=True)
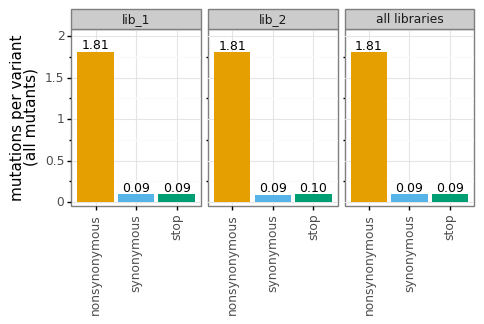
Or more silly:
[15]:
# NBVAL_IGNORE_OUTPUT
theme_set(theme_xkcd())
p = variants.plotNumCodonMutsByType(
"all", samples=None, heightscale=1.2, widthscale=1.2
)
_ = p.draw(show=True)
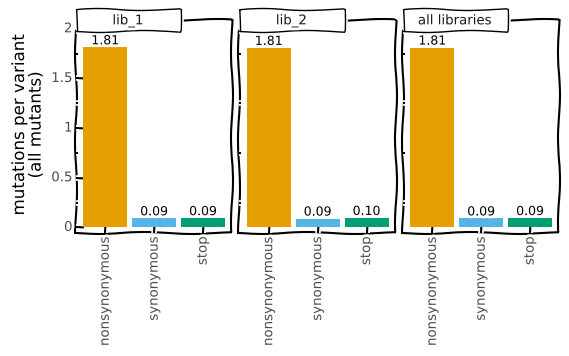
Note how the above call also used the heightscale and widthscale options (which exist for all plotting methods of a CodonVariantTable) to make the plot larger.
You can also set the orientation differently with orientation, and rename samples with sample_rename:
[16]:
# NBVAL_IGNORE_OUTPUT
theme_set(dms_variants.plotnine_themes.theme_graygrid()) # restore gray-grid theme
p = variants.plotNumCodonMutsByType(
"all",
samples="all",
orientation="v",
heightscale=1.2,
sample_rename={"loose_bottle": "loose bottle", "tight_bottle": "narrow bottle"},
)
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
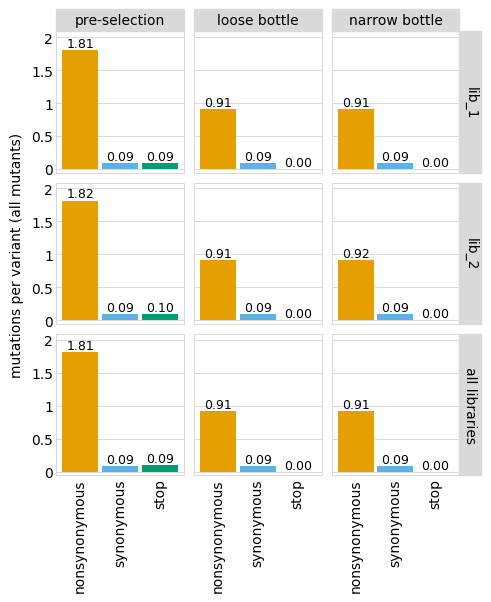
Or only show some of the facets. For instance, just show the individual libraries:
[17]:
# NBVAL_IGNORE_OUTPUT
p = variants.plotNumCodonMutsByType(
"all",
samples="all",
libraries=variants.libraries,
sample_rename={"loose_bottle": "loose bottle", "tight_bottle": "narrow bottle"},
)
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
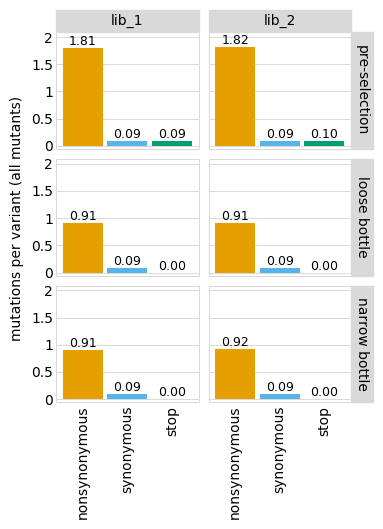
Note that if you just specify one library, by default the library name is not shown in the facet title:
[18]:
# NBVAL_IGNORE_OUTPUT
p = variants.plotNumCodonMutsByType(
"all",
samples="all",
libraries=["lib_1"],
sample_rename={"loose_bottle": "loose bottle", "tight_bottle": "narrow bottle"},
)
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
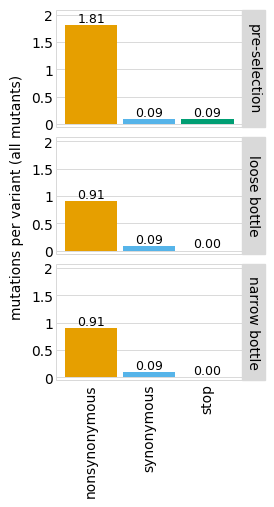
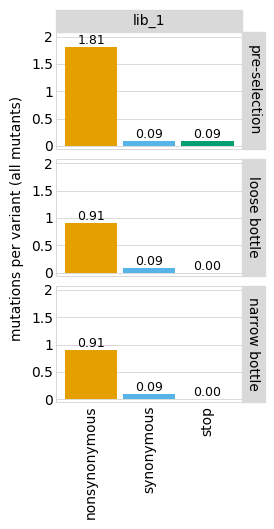
You can change this behavior by setting the one_lib_facet parameter to True:
[19]:
# NBVAL_IGNORE_OUTPUT
p = variants.plotNumCodonMutsByType(
"all",
samples="all",
libraries=["lib_1"],
sample_rename={"loose_bottle": "loose bottle", "tight_bottle": "narrow bottle"},
one_lib_facet=True,
)
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
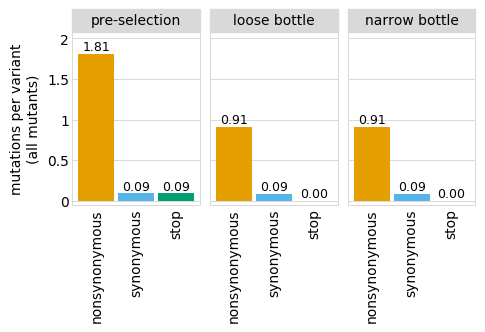
Or only the merge of all libraries:
[20]:
# NBVAL_IGNORE_OUTPUT
p = variants.plotNumCodonMutsByType(
"all",
samples="all",
libraries="all_only",
sample_rename={"loose_bottle": "loose bottle", "tight_bottle": "narrow bottle"},
orientation="v",
)
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
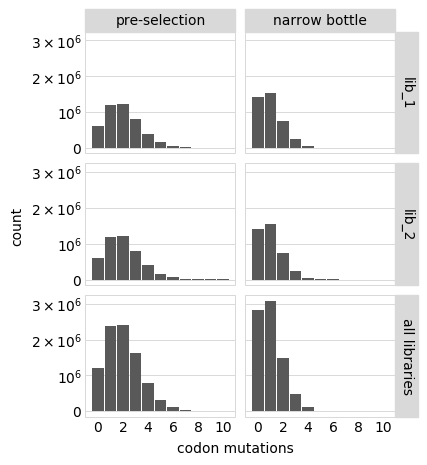
Or only show some samples:
[21]:
# NBVAL_IGNORE_OUTPUT
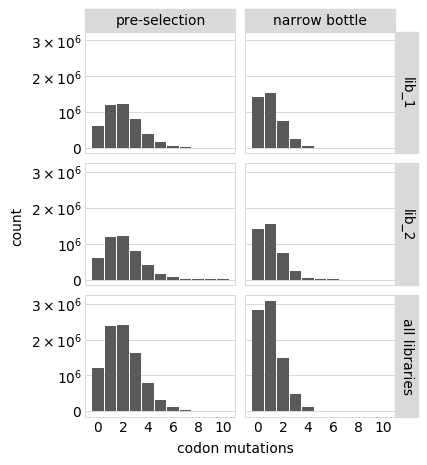
p = variants.plotNumMutsHistogram(
mut_type="codon",
samples=["pre-selection", "tight_bottle"],
orientation="v",
sample_rename={"loose_bottle": "loose bottle", "tight_bottle": "narrow bottle"},
)
p = p + theme(panel_grid_major_x=element_blank()) # no vertical grid lines
_ = p.draw(show=True)
You can also save the plots to image files using their save method. Here we show how to do this, saving the plot as a PNG to a temporary file and then displaying that PNG:
[22]:
with tempfile.NamedTemporaryFile(suffix=".png") as f:
p.save(f.name)
display(Image(f.name))